3rd Annual Meeting of the Boston Area Mitosis and Meiosis Community
Date:
My first poster presentation in the USA took place at the BAMM Meeting 2016 in Cambridge, MA. This is a local conference on cell division mechanisms. I was the only person who was doing computer modelling of biological system such as microtubule at the mesoscale.
My POSTER about the microtubule dynamics model:
Coarse-Grained Langevin Dynamics Modeling of Microtubules Assembly and Shortening.
E.Klyshko, K.A. Marx, F.I. Ataullakhanov, E.L. Grishchuk and V.Barsegov.
Abstract picture:
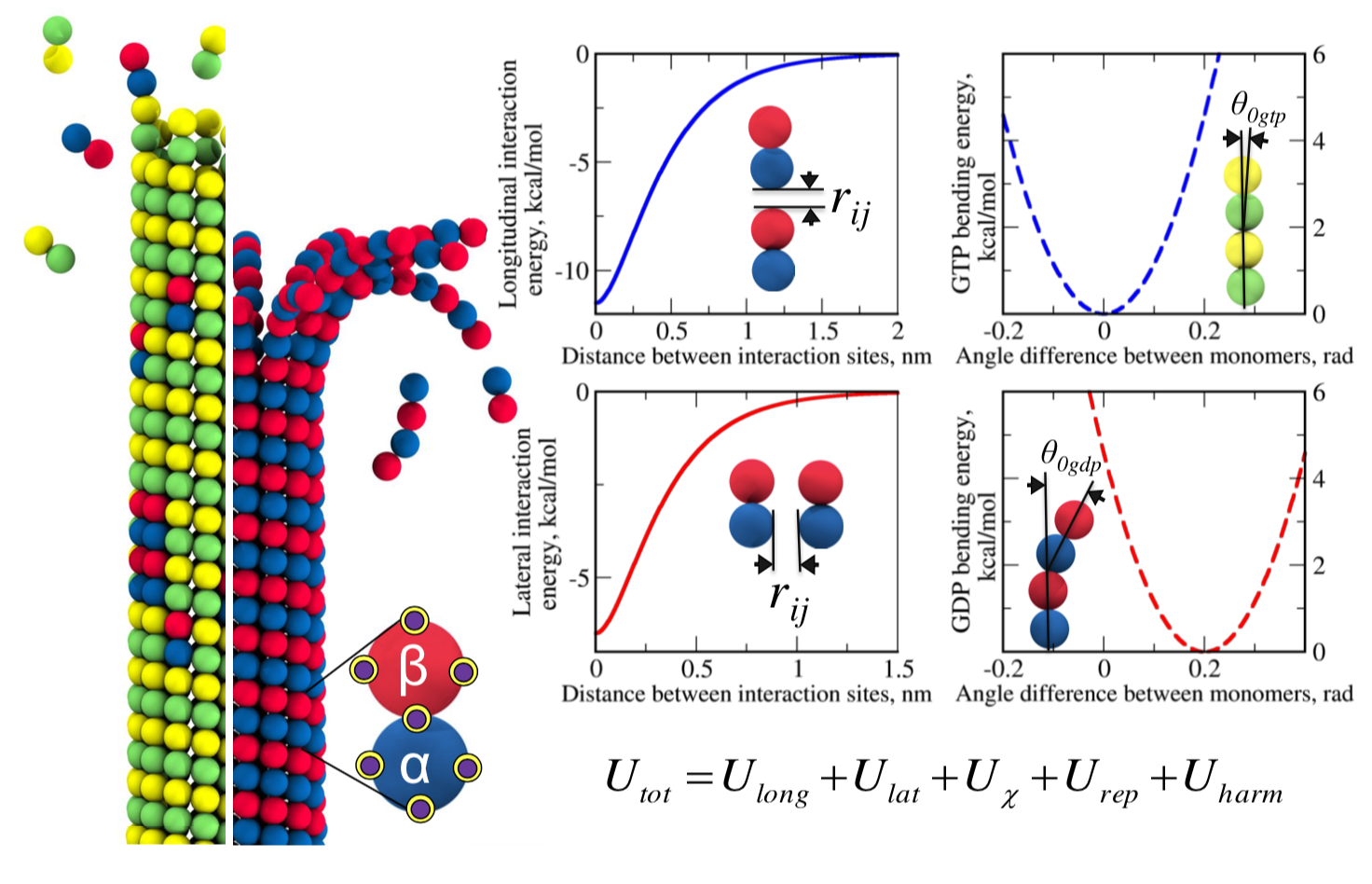
ABSTRACT:
We employed the multiscale computational modeling of microtubule (MT) polymers, which combine: i) all-atomic and Cα-based coarse-grained descriptions of α- and β-tubulin monomers; and ii) Langevin simulations of the bead-per- monomer model of the αβ-tubulin dimers, accelerated on Graphics Processing Units (GPUs), to characterize the kinetics and mechanisms of MTs’ assembly and shortening. In our model, the MT lattice is stabilized by the longitudinal and lateral non-covalent bonds between tubulin subunits; and the α- and β-tubulin monomers in dimers are allowed to bend around the MT cylinder axis and transverse directions. The MT protofilaments form curls in the right conformation consistent with recent experiments and splay outwards with correct bending rigidity and the GTP/GDP-bound tubulin-dependent equilibrium bending angle. Excellent agreement between the MT length vs. time profiles for the MT growth/shortening in vitro and in silico enabled us to obtain accurate model parameterization. The model offers a comprehensive quantitative platform to link the molecular tubulin characteristics with the physiological behavior of MT on multiple scales of length and time. The GPU-based implementation provides a powerful tool for the exploration of force-generating properties of MT polymers.
